Custom Genome Editing
Service Request FormNeed to generate mutant cell lines but too busy to do it yourself? ALSTEM offers complete genome editing services according to your specifications to help you accelerate your research. Empowered by CRISPR/Cas9 technology, our experts can generate genome edited cell lines carrying permanent knock-out, conditional knock-out, point mutation, and/or insertion to meet your needs.
Highlighted features of our genome editing service includes
- Diverse and personalized editing services with high standards: We provide service following your specifications to address your unique goals. Our experts have extensive experience in various types of editing, including permanent knock-out, conditional knock-out, point mutation, and insertion.
- Complete product pipeline: We design and clone your gRNA & HR donor plasmid, as well as generate stable single clonal cell lines for you. The turnaround time is ~2 to 5 months.
- Transparent manufacture and convenient communication: We provide timely and final project report to keep you updated and our scientists are always ready when you have questions.
- Comprehensive quality control: We validate your CRISPR cell lines with Sanger sequencing, as well as NGS if you prefer.
- Ready to use product: We provide 2 vials of cells at 0.5~1 X 106 cells/vial for each cell line we generate for you.
Please CONTACT US to discuss your gene editing project or fill in the form below and send it to us.
Background
Click to ReadGenome editing is a powerful tool to introduce additions, deletions, and substitutions in the genome of a living organism. The development of genome editing technologies involves with the use of meganucleases; zinc finger nucleases (ZFNs); transcription activator-like effector nucleases (TALENs); and, most recently, the CRISPR/Cas9 system (Figure 1).
- Meganucleases are nucleases with very long DNA-binding recognition sites which are from 14 to 40 nucleotides (Silva et al., 2011). Due to its highly specific and long recognition sequence, it is not applicable in modification on a target site in the genome.
- ZFN is an artificial restriction enzyme that is composed of 6 to 8 DNA recognition motifs, or zinc fingers, and the catalytic domain of DNA-cleaving enzyme, FokI. Each zinc finger recognizes 3 base pairs. The catalytic domain of Fok1 is then dimerized to induce double strand break at the target site of the genome.
- Similar to ZNFs, TALENs are composed of a DNA-cleaving protein fused with an array of nuclear effector domains. Each domain contains 32 to 34 amino acids which recognize a single base pair. The nuclear effector domains of two TALEN complexes are designed to bind to a specific sequence in the genome, which then allows dimerization of FokI, leading to double strand break at the target locus.
- CRISPR/Cas9 system is the most commonly used tool for genome editing currently. It has two components, a ~20 nucleotides small guide RNA (SgRNA) which binds to target DNA sequence and a Cas9 protein which exhibits nuclease function.
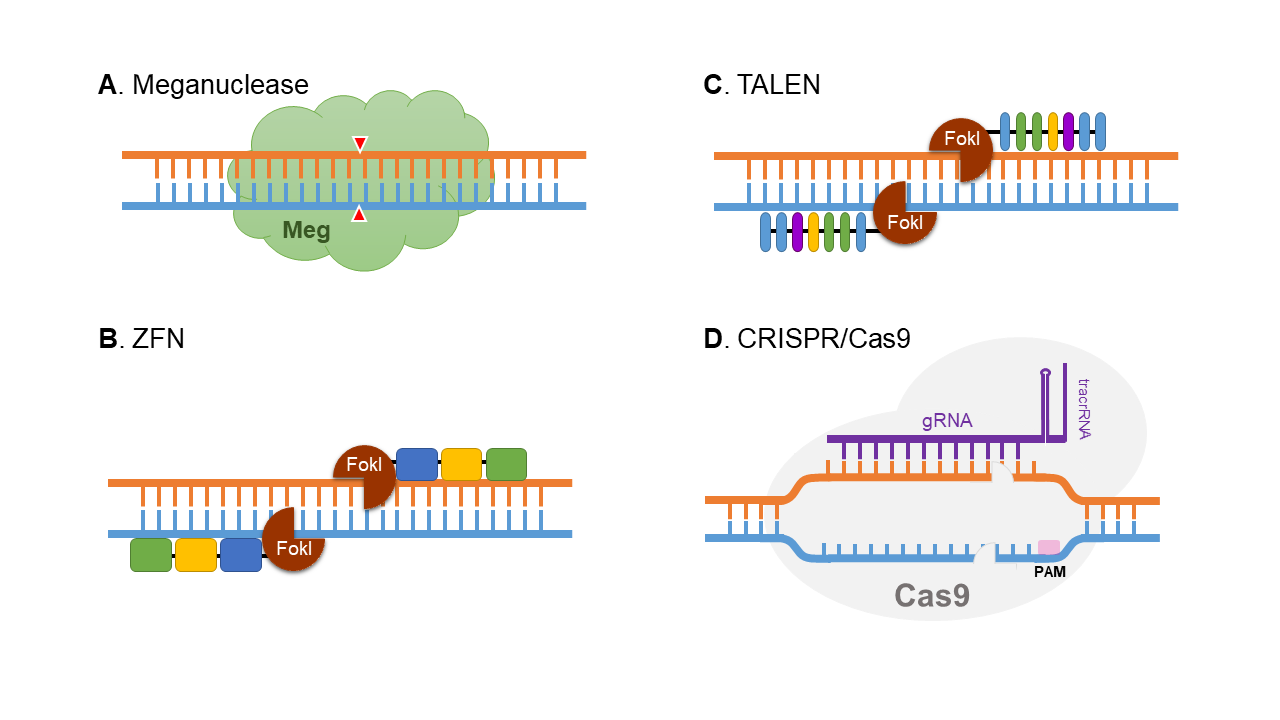
Figure 1. Schematic representation of four genome editing technologies. (A) Meganucleases are nucleases with long DNA recognition site, which might not be applicable for target-site editing. (B) ZFN combines restriction endonuclease and several zinc-finger-binding domains with each recognize 3 base pairs. Two ZFNs are designed to recognize the target sequence in the genome and FokI is dimerized to create double strand break. (C) TALEN is similar to ZFN with replacement of zinc-finger-binding domains with repeats of nuclear effector domains. Each nuclear effector domain recognizes single base pair. (D) CRISPR/Cas9 is a complex of artificial designed SgRNA and Cas9 nuclease. SgRNA binds to target DNA sequence which allows Cas9 to create double stranded break.
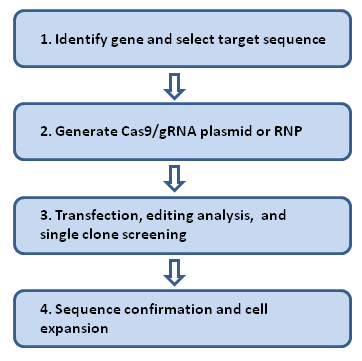
Our genome editing work flow
Case studies
-
Point Mutation
Human ApoE has three isoforms, ApoE2, ApoE3 and ApoE4. ApoE3 and ApoE4 differ from each another only at one amino acid residue at position 112. ApoE3, the common isoform, has Cystein (C) 112, whereas ApoE4 has Arginine (R) 112. ApoE4, a known major genetic risk factor for Alzheimer’s disease (AD), and researches has shown that it’s associated with early onset of AD in a gene dose-dependent manner. It may also contribute to age-related shrinking of the hippocampus and memory deficits in humans. To facilitate the studies of AD-related pathologies in neurons induced specifically by ApoE4, as well as for drug discovery, we developed ApoE3 cell line from human iPS ApoE4 cell line using CRISPR/Cas9. Cas9 protein/sgRNA ribonucleoprotein complexes (Cas9-RNPs) along with single-stranded oligo DNA nucleotides (ssODNs) containing R112C substitution were transfected into human iPS cells carrying human APOE4 gene. The cells were then expanded and single clones selected. As shown in the sequence data (Figure 2A), single-base substitution at amino acid position 112 was achieved.
A
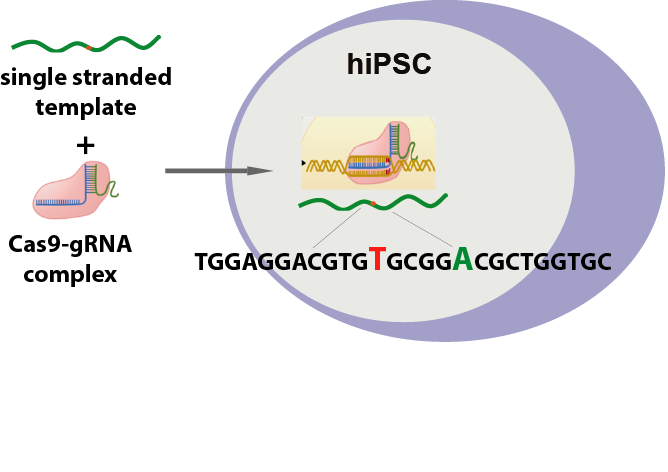
B
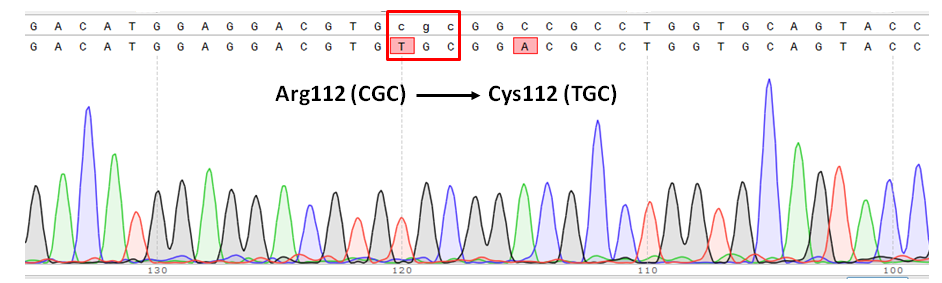
Figure 2. APOE homozygous point mutation. (A) Schematic representation of point mutation strategy. (B) Sequence confirmation of point mutation from ApoE4 to ApoE3 in human iPS cells. Red box indicates the desired point mutation.
-
Gene Knockout
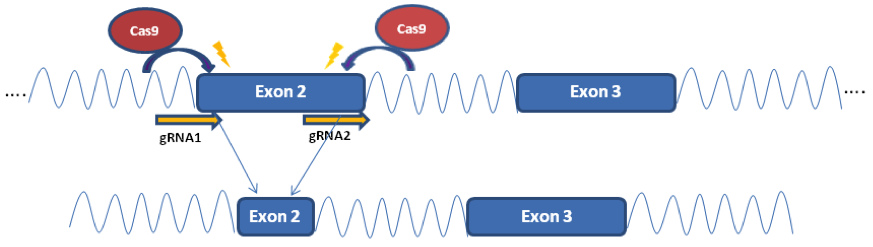
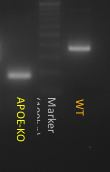
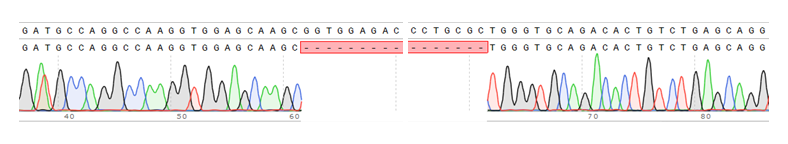
An apolipoprotein E (APOE) knockout (KO) cell line can be an important tool for functional study of APOE since APOE has been shown to play critical roles in lipid metabolism and neurodegenerative disease. Gene knockout is a potent and irreversible approach to inactivate a gene. A gene knockout may be achieved using the Nonhomologous end joining (NHEJ) after a double break of DNA at the chosen site using engineered endonucleases, such as TALEN and CRISPR/Cas9. Here in this example, we used CRISPR/Cas 9 system to generate ApoE KO cell line. A gRNA pair was designed to target Exon2 of APOE in human iPS cells, which is conserved among all 3 isoforms. This gRNA pair creates a deletion of 100 bp in Exon2, and cause frame shift and lead to early translation termination of APOE protein.
A
B
C
D
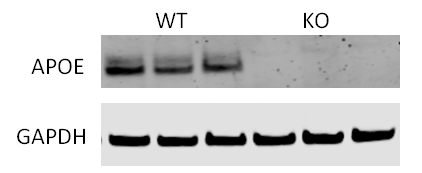
Figure 3. Generation of APOE KO cell line. (A) Schematic representation of gene knockout strategy. (B) Image of PCR amplification of genomic DNA from APOE KO clone and wild type line. (C & D) APOE KO confirmed by Sanger sequencing and western blotting results.
-
Gene Knockin
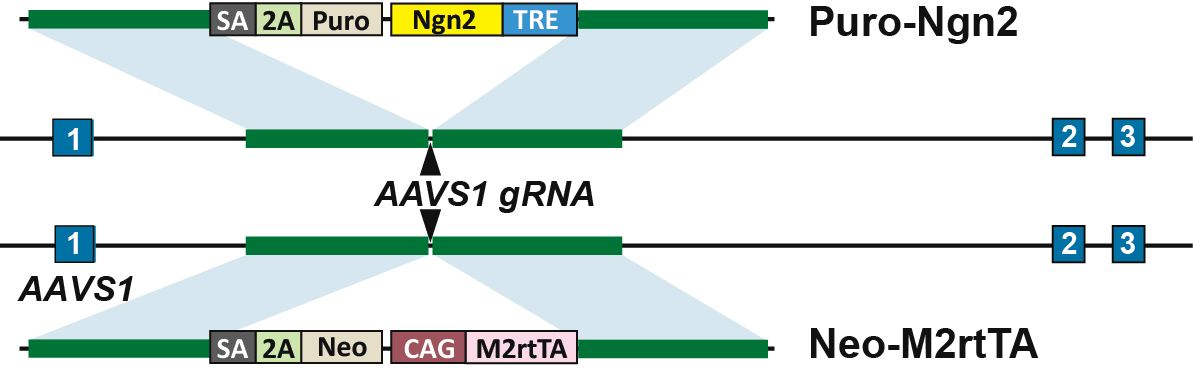
Neurogenin (Ngn) 2 is a basic helix-loop-helix (bHLH) transcription factor that promotes direct differentiation of pluripotent stem cells to functional excitatory neurons. In human genome, adeno-associated virus integration site 1 (AAVS1, also known as PPP1R12C) is a safe harbor for transgene integration, and it supports robust and sustained transgene expression. Precise insertion of tetracycline inducible Ngn2 into human iPS cells at AAVS1 site could generate a stable iPSC line for rapid neuronal differentiation upon induction.
Neurogenin (Ngn) 2 is a basic helix-loop-helix (bHLH) transcription factor that promotes direct differentiation of pluripotent stem cells to functional excitatory neurons. In human genome, adeno-associated virus integration site 1 (AAVS1, also known as PPP1R12C) is a safe harbor for transgene integration, and it supports robust and sustained transgene expression. Precise insertion of tetracycline inducible Ngn2 into human iPS cells at AAVS1 site could generate a stable iPSC line for rapid neuronal differentiation upon induction.
A
B
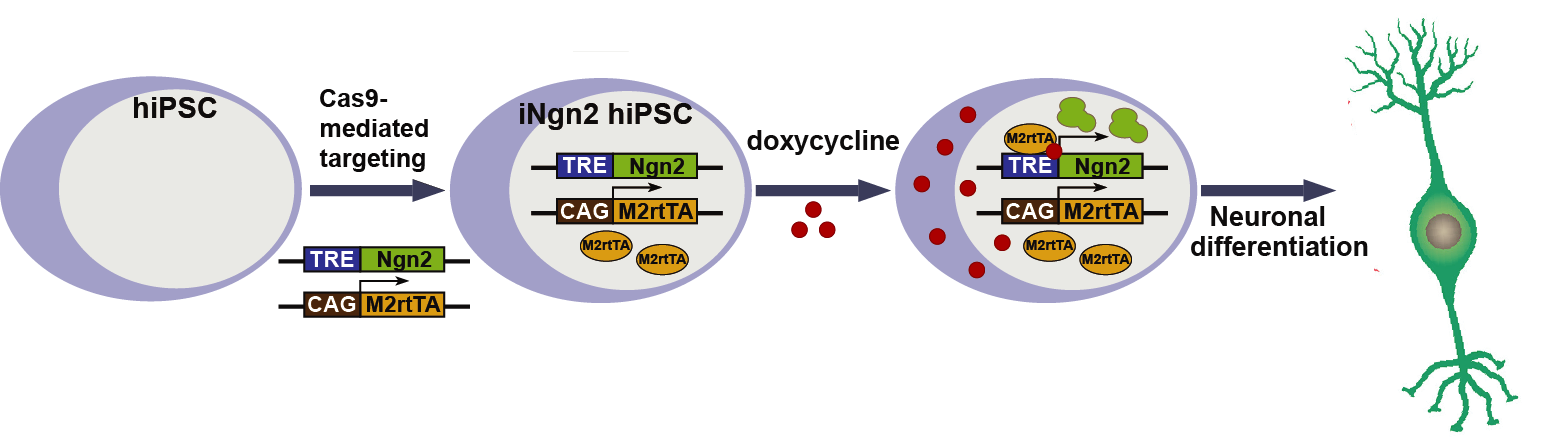
Figure 4. Knock in of Tet-on system into the AASV1 locus. (A) SgRNA was designed to target AASV1 locus. Ngn2 and M2rtTA were inserted at the AAVS1 locus through HDR pathway. M2rtTA constitutively expressed under the control of CAG promoter. (B) Upon Dox treatment, M2rtTA bound to TRE sequence which activated the expression of Ngn2. Human iPS cells were converted into excitatory neurons upon the expression of Ngn2.
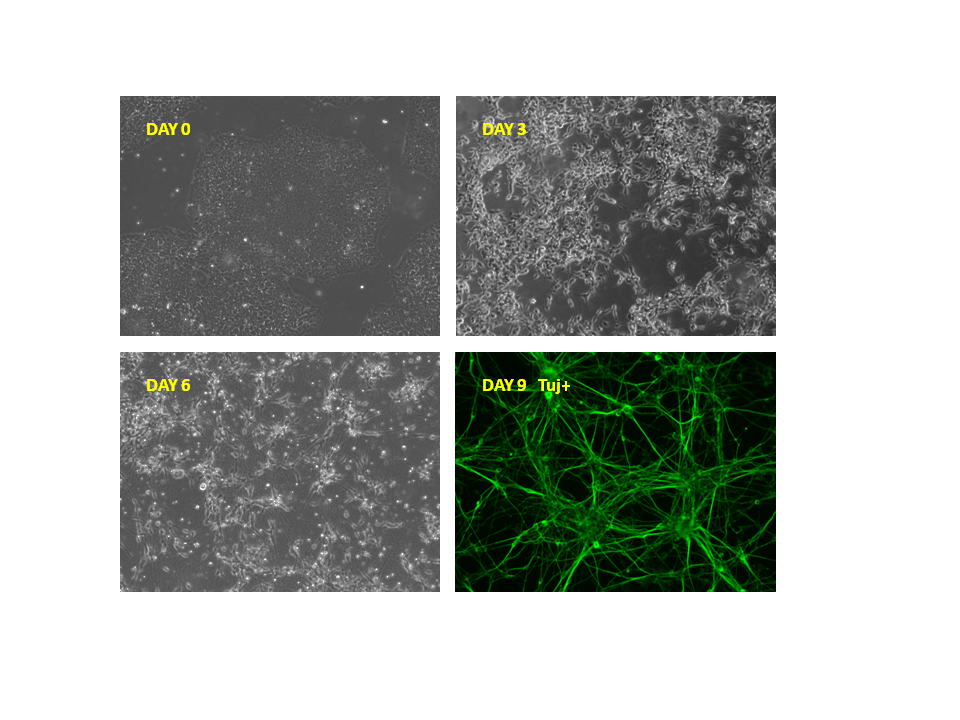
Figure 5. Upon doxycycline treatment, inducible Ngn2 iPS cells differentiated to neurons. (A) In the absence of doxycycline, the iPS cells retained the pluripotent state (B) 3 days after Dox treatment, iPS cells started to differentiate to neurons. (C) 6 days after Dox treatment, majority of cells had converted to neurons with extensive growth of axons and dendrites. (D) 9 days after Dox treatment, these cells had typical neuronal morphology and were beta-III-tubulin (Tuj) positive.
Publications
- Bertholet, AM., et al. H transport is an integral function of the mitochondrial ADP/ATP carrier. Nature (2019). 571, p515–520
- Diggins, NL., et al. α5β1 integrin trafficking and Rac activation are regulated by APPL1 in a Rab5-dependent manner to inhibit cell migration. Journal of Cell Science (2018) 131, jcs207019. doi:10.1242/jcs.207019
- Shahid, M., et al. Centromere protein F (CENPF), a microtubule binding protein, modulates cancer metabolism by regulating pyruvate kinase M2 phosphorylation signaling. Cell Cycle (2018). 17, 24, p2802–2818
- Wu, M., et al. Conditional gene knockout and reconstitution in human iPSCs with aninducible Cas9 system. Stem Cell Research (2018). 29, p6-14
